 Home Publications Links LEDCube
Home Publications Links LEDCube
|
This is my personal web page describing some of the optogenetics projects that I have worked on in the past years. Cellular (subcellular) Optogenetics Our work focusses on the development and application of cellular optogenetics (different from classical optogenetics, that uses light controlled ionchannels to control the activity of neurons). We utitilize plant light receptor domains to build light controllable versions of proteins that we are interested in. Such light controllable proteins can be turned off or on in a split second, and allow us to control proteins at the subcellular level. One of the light receptor domains that we utilize is called a LOV2 domain. This domain is part of a plant protein named phototropin, which in plants is essential for growth towards sunlight (phototropism). The LOV2 domain is particularly sensitive to blue light. 
State:
ready to load
loading ...
ready to play
playing
Load
Cancel
Play
Stop
Arrow indicates direction blue light (mixed vegetable sprouts) Here's a link to our recent review: "Lights, cytoskeleton, action: Optogenetic control of cell dynamics" In this review we discuss various strategies that have been employed to optogenetically control cytoskeleton/ cell dynamics. Current Opinion in Cell Biology, (includes free PMC full text).  fig 1 preview: comparison of various light receptors. A) A.s LOV2(phot1) structure in the dark(top) and lit(bottom) states B) A.t Cry1-PHR C) A.t PhyB-PSM D) structure of GFP for comparison. Below is a link to our paper on a blue light inactivated version of the microtubule plus-end-tracking protein EB1, which was published in Nature Cell Biology in 2018. EB1 is an important regulator of the microtubule cytoskeleton in eukaryotes. It regulates MT growth dynamics and mediates attachment of MTs to other subcellular structures, by recruiting over 40 different types of +TIPs to growing MT ends. We engineered a variant of EB1 which splits apart in the presence of blue light, and can no longer recruit these other +TIPs to the ends of MTs. This impacts MT growth, and MT attachment at the cell cortex. We engineered pi-EB1 by inserting a light sensitive protein module in the EB1 protein. This module consists of the plant light receptor domain LOV2, plus a a small protein, named Zdk1, that binds LOV2 in the dark but dissociates in the presence of blue light (LOVTRAP). Below you find a schematic of photo-inactivated(pi)-EB1, and images of a cell before and after inactivation of pi-EB1.  Figure 1. A light‐sensitive EB1 variant can replace endogenous EB1 function (click here for the full article). (a) Interaction of purified LOV2 domain and Zdk1 peptide analysed by native PAGE. Blue light results in dissociation of the LOV2/Zdk1 complex (arrow) that is upshifted compared to LOV2 or Zdk1. (b) Schematic of the photo‐inactivated π‐EB1 design resulting in reversible dissociation of the N‐terminal MT‐binding and the C‐terminal +TIP adapter domains upon blue light exposure. (c) Analysis of EB1 and π‐EB1 expression levels in H1299 cell lines in which π‐EB1 constructs were stably expressed and endogenous EB1 depleted by shRNA. Immunoblots were probed with antibodies specific to either the C‐ or N‐terminus of EB1. (d) Cell expressing Tubulin‐mCherry and EGFP‐tagged Zdk1‐EB1C before and after 5 s of exposure to 488 nm blue light resulting in dissociation of Zdk1‐EB1C from growing MT plus ends. (e) Analysis of the blue light‐induced EGFP‐Zdk1‐EB1C dissociation rate from MT ends. n = 9 cells. Error bars are 95% confidence intervals. Solid line is a fit with an exponential decay. Inset shows the comparison of dissociation half‐lives of Zdk1‐EB1C and SLAIN2, a +TIP that depends on EB1 for MT end association. (f) Cell expressing both halves of π‐EB1 fluorescently tagged showing that EB1NLOV2 remains on growing MT ends after blue light exposure. Time stamps indicate duration of blue light exposure. Dual‐wavelength images were acquired simultaneously using an emission image splitter. (g) Analysis of the amount of the two π‐EB1 halves bound to MT ends before and during 1 s of blue light exposure. In e and g, each symbol represents the average of 6‐8 measurements from one cell. p‐values were determined by Tukey‐Kramer HSD test. van Haren J, Charafeddine RA, Ettinger A, Wang H, Hahn KM, Wittmann T. Local control of intracellular microtubule dynamics by EB1 photodissociation. Nature Cell Biology, 2018 Mar 20; (3):252-261. doi: 10.1038/s41556-017-0028-5. Epub 2018 Jan 29. Nature Cell Biology, Article (click here) We also published supporting protocols on how to make cell lines expressing this optogenetic switch, and provide some advice on things to keep in mind when designing your own optoswitches using this "domain splitting" strategy. Wittmann T, van Haren J. Generation of cell lines with light-controlled microtubule dynamics. Protocol Exchange, 2018, published online 29 January 2018. doi:10.1038/protex.2017.155 Protocol Exchange, Protocols (click here) pi-EB1 constructs are available on Addgene (click here)  Practical implementation of optogenetics and live cell microscopy We recently published a chapter in
Here's a preview figure:  Here are two GIFs of the LED-cube in action 
State:
ready to load
loading ...
ready to play
playing
Load
Cancel
Play
Stop

State:
ready to load
loading ...
ready to play
playing
Load
Cancel
Play
Stop
Inactivation is almost complete at 300ms blue light pulses at 1 Hz In the near future i'll try to upload more detailed step-by-step instructions on how to assemble the LEDcube For more info also see: - Our work was highlighted on: the Addgene blog. - Mightex application note:"Spatial control of microtubule dynamics at the subcellular scale by patterned illumination" 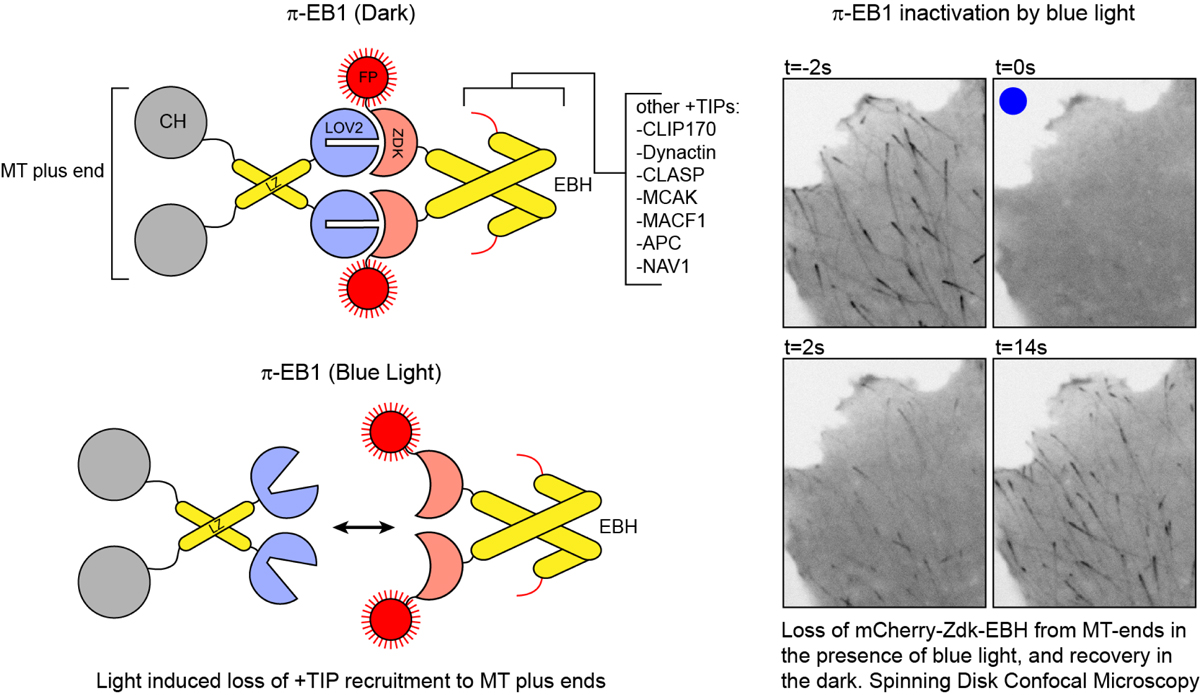 Schematic of pi-EB1(left) and example of pi-EB1 inactivation and reversion in a human cell |

|